Molecular microbial prospection for oil and gas and its preliminary application
-
摘要: 油气微生物勘探技术因其多解性小、信噪比高、受环境影响小、经济快速的特点,越来越受到勘探家的重视。随着高通量测序、分子生物学技术及生物信息学的快速发展,从微生物群落整体的角度描述特定油气指示微生物的变化,已成为未来微生物勘探技术的重要发展方向。介绍土壤样品采集、DNA提取与分析、判别模型建立和有利区预测等4个油气微生物分子勘探技术关键环节,并针对土壤DNA提取和油气指示微生物检测进行重点阐述。工业级DNA提取技术研究发现,Griffth方法提取并纯化土壤微生物DNA效果最佳。针对大样本量工程,使用高通量移液工作站和商用多孔板DNA抽提试剂盒可大幅提高勘探效率;综合稳定性同位素探针和高通量测序的分子检测技术研究探明了典型含油气区主要油气指示微生物菌种分布规律,高频油指示菌(AMNR族)为节杆菌、分枝杆菌、诺卡氏菌和红螺菌,高频气指示菌为甲基球菌、甲基杆菌、甲基孢囊菌。微生物分子勘探技术在鄂尔多斯盆地杭锦旗地区的初步应用表明,该技术在黄土塬地貌区油气识别精度显著提升,在含油气性评价方面效果良好,为鄂尔多斯盆地北部致密气有利区带筛选提供了参考依据。Abstract: Due to the advantages such as low multi-solution, high signal-to-noise ratio, low environmental impact, high efficiency and low cost, microbial prospection for oil and gas has been accepted for petroleum exploreration. With the rapid development of high-throughput sequencing, molecular biology and bioinformatics, describing the changes of specific oil and gas indicator micro-organisms from the perspective of microbial community as a whole has become an valuable progress of microbial exploration method for the future. In this paper, soil sampling, DNA extraction and analysis, discriminant model development and favorable area prediction are introduced, and soil DNA extraction as well as oil/gas indicator micro-organism detection are emphasized. With the study of industrial-grade DNA extraction technology, Griffth method has been considered as the best effect for the extraction and purification of soil microbial DNA. For projects with a large sample number, high-throughput pipetting workstations and commercial multi-well plate DNA extraction kits can greatly improve working efficiency. Integrated stable isotope probes and the molecular detection of high-throughput sequencing showed that the main hydrocarbon indicator microorganism strains typically imply the expected distribution. High frequency oil indicator bacteria (AMNR) are Arthrobacter, Mycobacterium, Nocardia, and Rhodospirillum, while high frequency gas indicator bacteria include Methylococcus, Methylobacterium, and Methylocystis. The preliminary application in the Hangjinqi area shows that the microbial molecular exploration technology can significantly improve the oil and gas identification accuracy in loess tableland regions, and has a good effect on the evaluation of oil and gas bearing property, which provides a reference for the selection of favorable tight gas regions in the northern part of Ordos Basin.
-
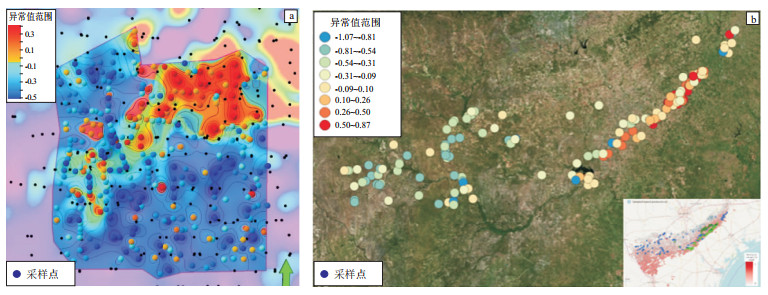
图 1 北美Haynesville地区(a)和Eagle Ford地区(b)页岩油气甜点预测准确率[12]
Figure 1. Prediction accuracy of shale oil and gas sweet spots in Haynesville (a) and Eagle Ford (b), USA
图 3 商用多孔板DNA抽提试剂盒抽提土壤DNA前后对比[12]
Figure 3. Soil DNA extraction with a commercial multiple well plate DNA extraction kit
表 1 土壤微生物总DNA提取方法
Table 1. Extraction methods of total DNA from soil microorganisms
表 2 典型含油气区主要油气指示微生物菌种
Table 2. Oil and gas indicator species in typical oil and gas areas
序号 区块 油品类型 地貌 主要油气指示菌种(丰度排列前四) 1 富安 凝析油 农田 Lysobacter、Methylomirabilis、Rhodospirillum、Nocardioides 2 长岭 凝析油 农田、草地 Rhodospirillum、Pseudonocardia、Arthrobacter、Mycobacterium 3 顺北 轻质油 沙漠 Nocardioides、Ralstonia、Pesudomonas、Methylobacillus 4 涠西 轻质油 海洋沉积物 Rhodobacter、Desulfococcus、Desulfuromonas、Desulfobulbus 5 江汉 稠油/轻质油 农田 Lysobacter、Rhodospirillum、Thermomonas、Streptomyces 6 春光 稠油/轻质油 沙漠化区 Nocardioides、Arthrobacter、Rhodospirillum、Streptomyces 7 南阳 稠油 农田 Rhodospirillum、Methylococcus、Arthrobacter、Nocardioides 8 镇泾 稠油 黄土塬 Nocardioides、Rhodospirillum、Arthrobacter、Aeromicrobium 9 沾化 稠油 农田 Arthrobacter、Pseudonocardia、Rhodospirillum、Methylomirabilis 10 玉北 稠油 沙漠 Pseudonocardia、Rhodospirillum、Amycolatopsis、Streptomyces 11 杭锦旗 天然气 草原 Methylocaldum、Methylococcus、Methylobacterium、Rhodospirillum 12 普光 天然气 山地 Methylococcus、Methylobacterium、Rhodospirillum、Nocardioides 13 焦石坝 天然气 山地 Methylococcus、Methylocystis、Methylobacterium、Rhodospirillum 14 涩北 天然气 盐碱地 Methylobacterium、Methylocystis、Nocardioides、Methylococcus 表 3 鄂尔多斯盆地杭锦旗新召地区土壤物理化学环境参数范围
Table 3. Range of soil physical and chemical environmental parameters in Xinzhao area, Hangjinqi, Ordos Basin
环境参数 均值 范围 含水量/% 7.1 5.0~10.6 pH 8.9 8.8~9.0 NOX-1-N/(μg·g-1) 0.96 0.51~1.30 50~100 μm颗粒 24.7 10.5~47.1 表 4 微生物异常与已知钻井含油气性吻合情况统计
Table 4. Statistics on the coincidence of microbial anomalies with known oil and gas content of wells
井号 综合异常分区 含油气性 化探异常符合情况
(符合率达55%)微生物异常符合情况
(符合率达61%)综合异常符合情况
(符合率达72%)锦25 背景区 气井 不符合 不符合 不符合 锦30 一级异常区 气井 符合 符合 符合 锦59 不确定区 气井 符合 符合 符合 锦60 二级异常区 气井 符合 符合 符合 锦61 不确定区 气井 符合 基本符合 符合 锦62 背景区 气井 不符合 不符合 不符合 锦63 背景区 气井 不符合 不符合 不符合 锦79 二级异常区 气井 符合 不符合 不符合 锦136 不确定区 气井 符合 不符合 基本符合 -
[1] 袁志华, 张玉清. 利用油气微生物勘探技术寻找页岩气有利目标区[J]. 地质通报, 2011, 30(2/3): 406-409. https://www.cnki.com.cn/Article/CJFDTOTAL-ZQYD2011Z1029.htmYUAN Zhihua, ZHANG Yuqing. Discussion of microbial prospecting of shale gas for favorable target areas[J]. Geological Bulletin of China, 2011, 30(2/3): 406-409. https://www.cnki.com.cn/Article/CJFDTOTAL-ZQYD2011Z1029.htm [2] 梅博文, 袁志华, 王修垣. 油气微生物勘探法[J]. 中国石油勘探, 2002, 7(3): 42-53. https://www.cnki.com.cn/Article/CJFDTOTAL-KTSY200203007.htmMEI Bowen, YUAN Zhihua, WANG Xiuyuan. Microbiological prospecting of oil and gas[J]. China Petroleum Exploration, 2002, 7(3): 42-53. https://www.cnki.com.cn/Article/CJFDTOTAL-KTSY200203007.htm [3] 汤玉平, 许科伟, 顾磊, 等. 油气微生物勘探理论与技术研究进展[J]. 石油实验地质, 2021, 43(2): 325-334. doi: 10.11781/sysydz202102325TANG Yuping, XU Kewei, GU Lei. et al. Recent progress in the theory and technology of microbial prospecting for oil and gas[J]. Petroleum Geology & Experiment, 2021, 43(2): 325-334. doi: 10.11781/sysydz202102325 [4] 顾磊, 许科伟, 汤玉平, 等. 基于高通量测序技术研究玉北油田上方微生物多样性[J]. 应用与环境生物学报, 2017, 23(2): 276-282. https://www.cnki.com.cn/Article/CJFDTOTAL-YYHS201702015.htmGU Lei, XU Kewei, TANG Yuping, et al. Microbial diversity in Yubei oil field determined by high-throughput sequencing[J]. Chinese Journal of Applied and Environmental Biology, 2017, 23(2): 276-282. https://www.cnki.com.cn/Article/CJFDTOTAL-YYHS201702015.htm [5] NING Zhuo, HE Ze, ZHANG Sheng, et al. Development of a prmA genes quantification technique and assessment of the technique's application potential for oil and gas reservoir exploration[J]. Energy Exploration & Exploitation, 2018, 36(5): 1172-1188. [6] DENG Yue, DENG Chunping, YANG Jinshui, et al. Novel butane-oxidizing bacteria and diversity of bmoX genes in Puguang gas field[J]. Frontiers in Microbiology, 2018, 9: 1576. doi: 10.3389/fmicb.2018.01576 [7] DENG Chunping, YU Xuejian, YANG Jinshui, et al. Universal indicators for oil and gas prospecting based on bacterial communities shaped by light-hydrocarbon microseepage in China[J]. Journal of Microbiology and Biotechnology, 2016, 26(7): 1320-1332. doi: 10.4014/jmb.1602.02045 [8] 顾磊, 许科伟, 汤玉平, 等. 基于高通量测序技术研究页岩气区上方微生物多样性[J]. 石油实验地质, 2020, 42(3): 443-450. doi: 10.11781/sysydz202003443GU Lei, XU Kewei, TANG Yuping, et al. Microbial diversity above a shale gas field using high-throughput sequencing[J]. Petroleum Geology & Experiment, 2020, 42(3): 443-450. doi: 10.11781/sysydz202003443 [9] 闫亮, 季苗, 贾宝迁, 等. 塔里木盆地顺北断溶体油气藏微生物特征及有利区预测[J]. 石油与天然气地质, 2020, 41(3): 576-585. https://www.cnki.com.cn/Article/CJFDTOTAL-SYYT202003014.htmYAN Liang, JI Miao, JIA Baoqian, et al. Microbial characteristics of Shunbei faulted-karst reservoirs and prediction of play fairways, Tarim Basin[J]. Oil & Gas Geology, 2020, 41(3): 576-585. https://www.cnki.com.cn/Article/CJFDTOTAL-SYYT202003014.htm [10] 闫亮, 贾宝迁, 季苗, 等. 塔里木盆地新和地区低幅度构造油气微生物特征及有利目标区预测[J]. 石油实验地质, 2020, 42(6): 1001-1008. doi: 10.11781/sysydz2020061001YAN Liang, JIA Baoqian, JI Miao, et al. Microbial characteristics of low-amplitude structures and prediction of favorable target areas in Xinhe area, Tarim Basin[J]. Petroleum Geology & Experiment, 2020, 42(6): 1001-1008. doi: 10.11781/sysydz2020061001 [11] 曹军, 周进松, 银晓, 等. 微生物地球化学勘探技术在黄土塬地貌区油气勘探中的应用[J]. 特种油气藏, 2020, 27(5): 53-60. https://www.cnki.com.cn/Article/CJFDTOTAL-TZCZ202005008.htmCAO Jun, ZHOU Jinsong, YIN Xiao, et al. Application of microbial geochemical exploration technology in oil and gas exploration in Loess Tableland Regions[J]. Special oil & Gas Reservoirs, 2020, 27(5): 53-60. https://www.cnki.com.cn/Article/CJFDTOTAL-TZCZ202005008.htm [12] TE STROET C, ZWAAN J, DE JAGER G, et al. Predicting sweet spots in shale plays by DNA fingerprinting and machine learning[C]//SPE Unconventional Resources Technology Conference. Austin: SPE, 2017. [13] CLEGG C D, RITZ K, GRIFFITHS B S. Direct extraction of microbial community DNA from humified upland soils[J]. Letters in Applied Microbiology, 1997, 25(1): 30-33. [14] DEVI S G, FATHIMA A A, RADHA S, et al. A rapid and economical method for efficient DNA extraction from diverse soils suitable for metagenomic applications[J]. PLoS One, 2015, 10(7): e0132441. [15] YAN Tingfen, YE Qi, ZHOU Jizhong, et al. Diversity of functional genes for methanotrophs in sediments associated with gas hydrates and hydrocarbon seeps in the Gulf of Mexico[J]. FEMS Microbiology Ecology, 2006, 57(2): 251-259. [16] 顾磊, 梅泽, 许科伟, 等. 气态烃诱导下油气微生物数量及功能基因变化特征[J]. 微生物学报, 2017, 57(1): 43-53. https://www.cnki.com.cn/Article/CJFDTOTAL-WSXB201701007.htmGU Lei, MEI Ze, XU Kewei, et al. Changes of microbial abundance and functional genes in oil and gas under gaseous hydrocarbon condition[J]. Acta Microbiologica Sinica, 2017, 57(1): 43-53. https://www.cnki.com.cn/Article/CJFDTOTAL-WSXB201701007.htm [17] 丁力, 郝纯, 吴宇兵, 等. 微生物油气检测技术在准噶尔盆地油气勘探中应用[J]. 中国石油勘探, 2021, 26(3): 136-146. https://www.cnki.com.cn/Article/CJFDTOTAL-KTSY202103013.htmDING Li, HAO Chun, WU Yubing, et al. Progress and application of microbial oil and gas detection technology in oil and gas exploration, Junggar Basin[J]. China Petroleum Exploration, 2021, 26(3): 136-146. https://www.cnki.com.cn/Article/CJFDTOTAL-KTSY202103013.htm [18] ZHANG Cuiyun, HE Ze, ZHANG Sheng, et al. A DNA-based analysis of a microbial technique for the prospecting of oil and gas applied to a known oil field, China[J]. Geomicrobiology Journal, 2017, 34(1): 63-70. -





 下载:
下载:







 苏公网安备32021102000780号
苏公网安备32021102000780号